Abstract
Fungal metabolic volatiles attract Drosophila suzukii which oviposits in ripening fruits, but there are few data describing the fungal microbiomes of commercial fruits susceptible to this insect pest. We tested the hypothesis that fruit type and ripening stage have a significant effect on fruit surface fungal communities using DNA metabarcoding approaches and found strong support for differences in all three fungal community biodiversity metrics analysed (numbers, types, and abundances of taxa). There was an average fivefold greater difference in fungal communities between sites with different fruit types (strawberry, cherry, raspberry, and blueberry) than across fruit developmental stages, demonstrating site and/or fruit type is the greater factor defining fungal community assemblage. The addition of a fungal internal standard (Plectosphaerella cucumerina) showed cherry had relatively static fungal populations across ripening. Raspberry had a greater prevalence of Saccharomycetales yeasts attractive to D. suzukii, including Hanseniaspora uvarum, which aligns with reports that raspberry is among the fruits with greatest susceptibility and attraction to D. suzukii. Greater knowledge of how yeast communities change during fruit maturation and between species or sites may be valuable for developing methods to manipulate fruit microbiomes for use in integrated pest management strategies to control D. suzukii.
Introduction
Fungi are widespread in the environment and are important components of agricultural and natural ecosystems where they play key roles in nutrient turnover. Fruit associated fungi may have both positive and negative impacts on the quality of products derived from horticultural systems by causing spoilage1 or beneficial attributes in fermented beverages such as wine2. Fruit surfaces are home to complex and dynamic microbial communities which are affected by a number of factors including fruit species3,4 and variety5,6, ripening stage7,8,9, plant organ9, geographic location6,10,11,12 and farming practices13,14. Fruit fungal communities are dominated by the Ascomycota and Basidiomycota phyla, with Ascomycota comprising 52–97% and Basidiomycota 4–24% of species on a range of fruits including Vitis vinifera (grape)12, Malus pumila Mill. (apples), Ribes nigrum (blackcurrants)4, and Fragaria × ananassa (strawberries)9.
Drosophila are saprotrophic and thus dependent on microbes for nutrition; complex interactions between Drosophila, microbes and fruit have been described15. Unlike most other Drosophila species, Drosophila suzukii is able to oviposit in ripening fruit due to a morphologically modified ovipositor16. Drosophila suzukii causes economic losses through direct fruit damage by ovipositing and subsequent larval feeding, including indirect damage caused by secondary infection from microbes via wounds as entry points17,18,19. The economic damage caused by D. suzukii is significant, with losses estimated at $511.3 million in just three USA states in 200820. Recent geographic range expansion including invasions into the USA and mainland Europe in 2008 and the UK in 201220,21,22 have resulted in D. suzukii now significantly threatening soft and stone fruit production in most Northern Hemisphere temperate regions. Yeasts are an important source of nutrients for D. suzukii as they provide a protein source important for egg development23. Female D. suzukii prefer to oviposit on yeast-colonised fruits24, and female fecundity, larva development and survival is affected by yeast species25,26. It is increasingly documented that D. suzukii are attracted to volatile chemicals produced by budding yeasts in the Saccharomycetales order, including Hanseniaspora uvarum, Hanseniaspora opuntiae, Saccharomyces cerevisiae, Metschnikowia pulcherrima, Candida zemplinina, Candida californica, Pichia terricola and Pichia pijperi27,28,29,30,31. In addition to single species, we have recently shown that various combinations of yeasts involving C. zemplinina, P. pijperi, M. pulcherrima and H. uvarum are attractive to D. suzukii30. Moreover, there is an overlap between Saccharomycetales yeasts found on cherry and raspberry and those in D. suzukii guts, particularly Hanseniaspora species32,33. There is also recent evidence to show reduced olfactory attraction to raspberries when they are infected with Botrytis cinerea, a fruit fungal pathogen34. These observations can be exploited for the control of D. suzukii in various traps and baits15. Thus, there is value in understanding the general communities of fungi and especially the communities of Saccharomycetales naturally associated with various fruits as they ripen as these may modulate the attraction of D. suzukii.
Fungi associated with crops and foods were originally evaluated by culture-based approaches, but work shows that up to 95% of fungi on fruits may be missed using these methods12. The PCR amplification of specific diagnostic ‘barcode’ areas from DNA which has been directly extracted from substrates of interest may circumvent this non-culturable issue12. DNA barcode metagenomics studies have reported significant differences between fungal communities on the surfaces of apples and blackcurrants4, as well as between Hippophae rhamnoides L. (sea buckthorn), Aronia melanocarpa (black chokeberry), and Ribes rubrum (red and white currants)3. Further, there are reports that fungal communities differ between varieties within fruit species in both the Northern and Southern Hemisphere, e.g. between Chardonnay and Syrah grape varieties in New Zealand6 and Zinfandel, Chardonnay and Cabernet Sauvignon in California10. One of the drawbacks of barcode amplicon sequencing (and metagenomic analyses generally) is that it only allows the relative abundances of taxa to be analysed, and the underlying absolute biological abundances of taxa are not known unless controls for abundances are included. Various metagenomic studies have attempted to quantify absolute abundances by the use of internal standards35, but to our knowledge no previous work has attempted this for fungal communities on fruit. In addition, there are few data characterising changes in fruit fungal communities through ripening generally. Studies on table grapes and sea buckthorn berries suggested these microbial communities changed over time8,9,36. Abdelfattah et al.9 observed significant differences in fungal community structure between immature and mature strawberry fruit with unweighted UniFrac analysis (P = 0.003), but communities were dominated by Botrytis and Cladosporium genera, suggesting the difference across ripening was driven by a subtle shift in rare taxa.
Data on the microbial composition of fruits and their maturation-related changes is generally limited. We are not aware of any studies that have comprehensively identified the fungal and yeast communities associated with different ripening stages of commercially important fruit which are susceptible to D. suzukii. To help fill this knowledge gap, we investigated the general fungal and Saccharomycetales (budding yeasts) communities on blueberry, cherry, raspberry, and strawberry during ripening in the UK, using a barcode metagenomics approach. Further, we aimed to do this quantitatively by spiking samples with a known number of Plectosphaerella cucumerina cells as an internal standard. We test the hypothesis that both fruit type and ripening stage have a significant effect on general fungal and Saccharomycetales yeast communities and evaluate whether there are differences in specific yeasts known to be attractive to D. suzukii.
Results
Six biological replicates each were sampled from four fruit species (blueberries, cherries, raspberries, and strawberries) at four developmental stages. Developmental stages were based on fruit pigmentation ranging from unripe (green) to fully ripe (red/purple/navy; Fig. S1) throughout June to September in 2018. Ten fruits (except blueberries N = 20) were collected for each species per replicate, and this was replicated six times for each ripening stage for each fruit at different sites.
Quantitative analysis of fungal communities
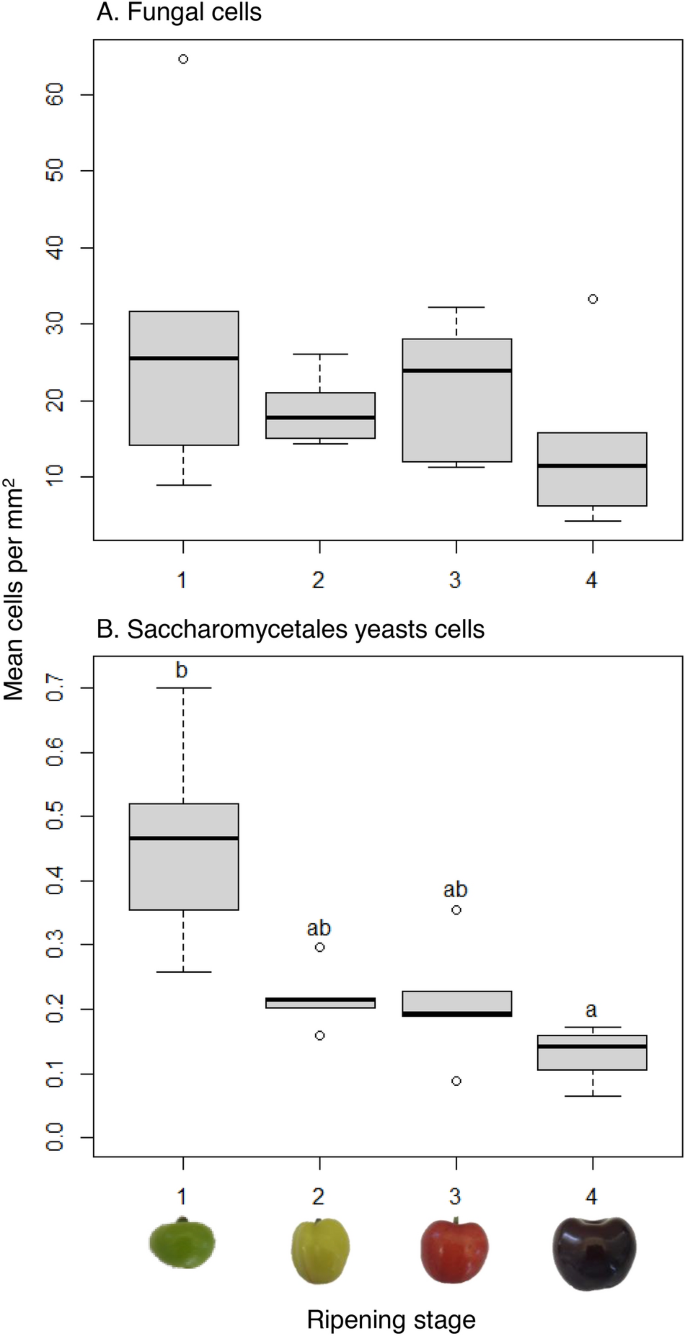
Metabarcoding analysis is generally not quantitative, but the addition of 265 P. cucumerina cells to sub-samples prior to DNA extraction served as an internal standard to attempt an estimation of the size of fungal populations. One replicate spiked with the internal standard of the strawberry stage 3 samples was removed due to poor sequence quality leaving 96 non-spiked and 95 spiked samples which produced a total of 38,445,395 reads that clustered into 1712 > 97% identity Amplicon Sequence Variants (ASV), which from here-in we call phylotypes (Table S1). Blast searches across all phylotypes for matches to the P. cucumerina internal standard’s ITS sequence generated from Sanger sequencing revealed one phylotype that matched with 100% identity. Plectosphaerella cucumerina was naturally present in 21 of the 95 non-spiked samples and comprised of a total of 444 reads. Cherry was the only fruit where the internal standard was reliably recovered: 23 of 24 spiked samples and only one of 24 non-spiked samples contained the internal standard phylotype. After internal standard DNA read normalisation, the mean (± SE) number of fungal cells from each of the useable 23 pairs of cherry replicates was 307,323 (± 39,090) cells. The range of phylotype cell abundance across all cherry samples was 3.9 million for an Aureobasidium phylotype to 3 cells for a phylotype taxonomically assigned no higher level than kingdom. There was no significant change in total fungal cell numbers across cherry maturation stage (Kruskal–Wallis, chi-squared = 2.63, P = 0.45; Fig. S2), but fruit surface areas also increased significantly (Kruskal–Wallis, chi-squared = 19.70, P = 0.0002, Fig. S2). When cell numbers were normalised for surface area this revealed that absolute fungal population sizes remained static across cherry maturation stages (Kruskal–Wallis, chi-squared = 2.49, P = 0.48; Fig. 1A). However, there was a significant change in absolute Saccharomycetales cell numbers when normalised for cherry surface area across maturation (Kruskal–Wallis, chi-squared = 15.30, P = 0.002): stage 1 had significantly greater absolute Saccharomycetales cell numbers than stage 4 (P = 0.0007; Fig. 1B). Six individual Saccharomycetales yeast phylotypes from the genera Debaryomyces, Saccharomyces, Kodamaea, one from the family Pichiaceae, and phylotypes with > 97% homology to M. pulcherrima and Metschnikowia gruessii, had significantly greater abundances on ripening stage 1 than 4 (P values span 0.045 to 0.006).
Absolute fungal cell abundances on cherry epicarp. Number of total fungal (A) and Saccharomycetales yeasts (B) cells per mm2 of cherry epicarp (N = 6 except, stage 3 and 4, N = 5) at four ripening stages (1, unripe/green fruit; 2, de-greening fruit; 3, ripening fruit; and 4, fully ripe/harvest fruit) estimated from DNA read abundances normalised to DNA abundances from the deliberate addition of 265 live Plectosphaerella cucumerina cells prior to DNA extraction. Different lower-case letters above bars show significant differences between ripening stages at P > 0.05, Dunn’s comparisons post-hoc with Benjamini–Hochberg multiple comparison correction.
Overview of fungal diversity across all fruit samples
The P. cucumerina internal standard phylotype was removed from all samples, and the sequence data normalised and analysed. A total of 1712 fungal phylotypes was revealed, comprising seven phyla, 25 classes, 96 orders, 197 families, and 280 genera. The most abundant and diverse phylum was Ascomycota, comprising 92.2% of reads and 57.3% of phylotypes, followed by Basidiomycota (7.7% reads and 33.6% phylotypes), Zygomycota (0.1% and 1.1%), Chytridiomycota (> 0.1% and 0.7%), Mucoromycota (> 0.1% and 0.3%), Glomeromycota and Rozellomycota (both > 0.1% and 0.1%; Fig. S3A). A phylotype from the Cladosporium genus was the most common phylotype across all samples, comprising 60.8% of reads. A total of 87 phylotypes from the order Saccharomycetales (budding yeasts) was detected, comprising 1,792,782 DNA reads (4.7% of the total) spanning 10 families and 25 genera. Metschnikowia was the most abundant Saccharomycetales genus (40.0% of Saccharomycetales reads), followed by Hanseniaspora (38.2%), then Pichia (5.2%), with the remaining genera contributing fewer than 3% each. Candida was the most diverse genus within the order Saccharomycetales accounting for 21.8% of phylotypes, despite only comprising 2.4% of reads, followed by Metschnikowia (11.5%), Hanseniaspora (8.0%) and Pichia (6.9%), with each of the remaining genera contributing fewer than 3.5% of phylotypes each (Fig. S3B). The most common Saccharomycetales yeast across all samples was a phylotype from the genus Hanseniaspora with > 97% homology to H. uvarum and comprised 38.2% of the total Saccharomycetales reads (Fig. S3B).
The effect of fruit species and ripening stage on epicarp fungal communities
We analysed differences in three biodiversity metrics to evaluate the effect of fruit species and maturation stage on fungal communities: differences in the absolute numbers of phylotypes (richness); differences in the types of phylotypes (i.e. presences/absences); and differences in the relative abundances of phylotypes (community composition) following Morrison-Whittle et al.14 and Morrison‐Whittle and Goddard37.
Fungal phylotype richness
Phylotype richness was not normally distributed (Shapiro-Wilks, P = 0.008) but square root transformation allowed the data to conform to the assumptions of ANOVA. There was a significant effect of both fruit type and ripening stage on the number of fungal phylotypes, including an interaction between the two (F3,175 = 18.58, P = 1.65 × 10–10; F3,175 = 5.00, P = 0.002 and F9,175 = 6.69, P = 3.25 × 10–8 respectively). Comparisons of effect sizes revealed fruit type (ω2 = 0.30) had a 4.4 times greater effect than ripening stage (ω2 = 0.068) on fungal phylotype richness. Disregarding ripening stage, cherry (mean ± SE number of phylotypes = 98 ± 4.1) had significantly more fungal phylotypes than blueberry (68 ± 3.7), raspberry (72 ± 2.9) and strawberry (76 ± 3.2) (Tukey’s HSD, P < 1.0 × 10–7, P = 2.0 × 10–7 and P = 2.56 × 10–5 respectively), which did not differ from one another (Fig. S4). Disregarding fruit type, ripening stage 2 (mean ± SE number of phylotypes = 85 ± 2.9) and 3 (82 ± 4.1) had significantly more fungal phylotypes than stage 1 (P = 0.001 and P = 0.033, respectively), but numbers at stages 1 and 4 were not significantly different (Fig. S4). The absolute time points for sampling did however differ between fruits due to different maturation timings.
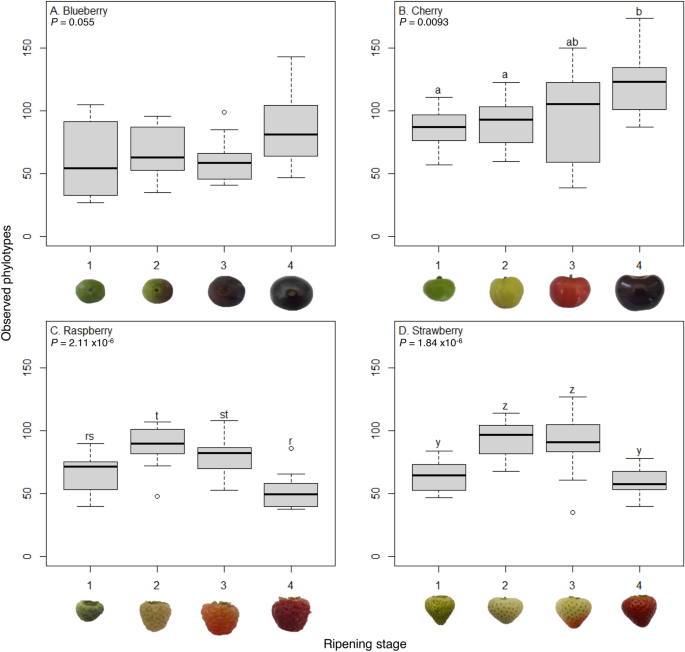
As there was a significant interaction between fruit and ripeness stage, we investigated the effect of ripening stage on each fruit separately. All data residuals were normally distributed (Shapiro–Wilks, P > 0.05) and there was a significant effect of ripening stage on the number of fungal phylotypes for cherry, raspberry, and strawberry (one-way ANOVA: F3,44 = 4.33, P = 0.0093; F3,44 = 13.56, P = 2.11 × 10–6 and F3,44 = 13.86, P = 1.84 × 10–6, respectively, Fig. 2), but not blueberry (F3,44 = 2.27, P = 0.055). On cherries phylotype numbers increased during ripening, but raspberry and strawberry had greater numbers at intermediate stages of fruit maturation (Fig. 2).
Number of observed phylotypes across fruit types and maturation stages. Number of fungal phylotypes across four ripening stages (1, unripe/green fruit; 2, de-greening fruit; 3, ripening fruit; and 4, fully ripe/harvest fruit) for blueberry, cherry, raspberry and strawberry (N = 12 except N = 11 for strawberry stage 3). Numbers of fungal phylotypes differ across ripening stages for cherry, raspberry and strawberry but not blueberry (ANOVA, P values shown). Where significant, different lowercase letters represent significant differences in phylotype numbers within each fruit (P < 0.028) with separate Dunn’s comparisons post-hoc (with Benjamini–Hochberg multiple comparison correction). Different letter groups show any significant differences between ripening stages within each fruit separately.
There was a significant effect of fruit type but not ripening stage on the number of Saccharomycetales budding yeast phylotypes (Kruskal–Wallis, chi-squared = 75.66, df = 3, P = 2.61 × 10–16 and chi-squared = 5.50, df = 3, P = 0.14 respectively). Raspberry (mean ± SE number of phylotypes = 12 ± 0.60) harboured significantly more Saccharomycetales phylotypes than strawberry (10 ± 0.74), cherry (7 ± 0.70), and blueberry (4 ± 0.31; Tukey’s HSD, P = 0.044, P = 2.9 × 10–6 and P = 1.5 × 10–15 respectively). Strawberry harboured significantly more phylotypes than cherry and blueberry (Tukey’s HSD, P = 0.007 and P = 2.6 × 10–9), and cherry harboured significantly more than blueberry (Tukey’s HSD, P = 0.001) (Fig. S5). Both Shannon’s and Simpson’s diversity indexes, which analyse the distribution of phylotype abundances, revealed differences between fruit species and ripening stage in line with the above findings (Table S2).
Presence/absence of fungal phylotypes
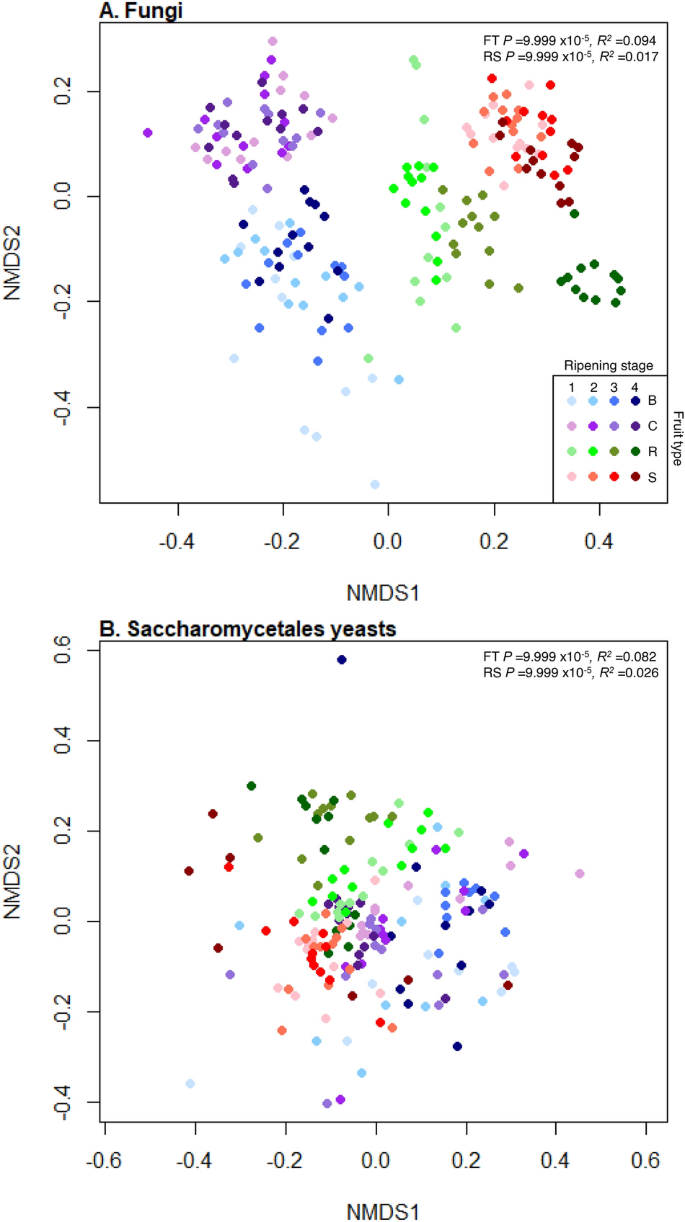
Both fruit type and ripening stage significantly influenced the types of fungi present (PermANOVA, R2 = 0.094, P = 9.999 × 10–5 and R2 = 0.017, P = 9.999 × 10–5, respectively, Fig. 3A) and there was a significant interaction between fruit type and ripening stage (R2 = 0.013, P = 9.999 × 10–5). Comparisons of effect sizes (R2 values) showed fruit type had approximately 5.5 greater influence than ripening stage on the types of fungal phylotypes present. As there was a significant interaction between fruit and ripening stage, the effect of ripening stage on fungal communities was investigated for each fruit separately. Ripening stage significantly influenced the types of fungal phylotypes present on all fruit (blueberry R2 = 0.043, 9.999 × 10–5; cherry R2 = 0.060, P = 9.999 × 10–5; raspberry R2 = 0.13, P = 9.999 × 10–5 and strawberry R2 = 0.055, P = 9.999 × 10–5, Fig. S6). There were significant differences in presences of fungal phylotypes between all fruits and ripening stages (post-hoc pairwise PermANOVAs: P = 9.999 × 10–5, R2 range 0.09–0.20; Fig. 3A; Supplemental Tables S3, S4).
NMDS plots representing the differential presences of fungal phylotypes. Nonmetric Multidimensional Scaling (NMDS) plots of binary Jaccard measures of community dissimilarity of (A) total fungal communities and (B) Saccharomycetales budding yeasts on blueberry (blue), cherry (purple), raspberry (green) and strawberry (red) at four ripening stages (1, unripe/green fruit; 2, de-greening fruit; 3, ripening fruit; and 4, fully ripe/harvest fruit; denoted by shade of colour, lightest shade for green fruit and moving through to darkest shade for fully ripe/harvest). Both total fungal and Saccharomycetales yeasts communities significantly differ in the presences of phylotypes across all fruit types (FT) and ripening stages (RS) by PermANOVA (values shown top right).
Both fruit type and ripening stage significantly influenced the types of Saccharomycetales phylotypes present (PermANOVA, R2 = 0.082, P = 9.999 × 10–5 and R2 = 0.026, P = 9.999 × 10–5, respectively, Fig. 3B) with a significant interaction between fruit type and ripening stage (R2 = 0.024, P = 9.999 × 10–5). In line with the general fungal community, comparisons of R2 values showed fruit type had approximately 3.15 times greater effect than ripening stage on the Saccharomycetales phylotypes present. Ripening stage significantly influenced the types of Saccharomycetales phylotypes on each fruit separately (blueberry R2 = 0.065, P = 0.0008; cherry R2 = 0.080, P = 0.0004; raspberry R2 = 0.27, P = 9.999 × 10–5 and strawberry R2 = 0.084, P = 9.999 × 10–5). There were significant differences in presences of different Saccharomycetales yeast phylotypes between all fruits and ripening stages (post-hoc pairwise PermANOVAs: P = 9.999 × 10–5, R2 range 0.06–0.15; Supplemental Tables S5, S6).
Relative abundances of fungal phylotypes
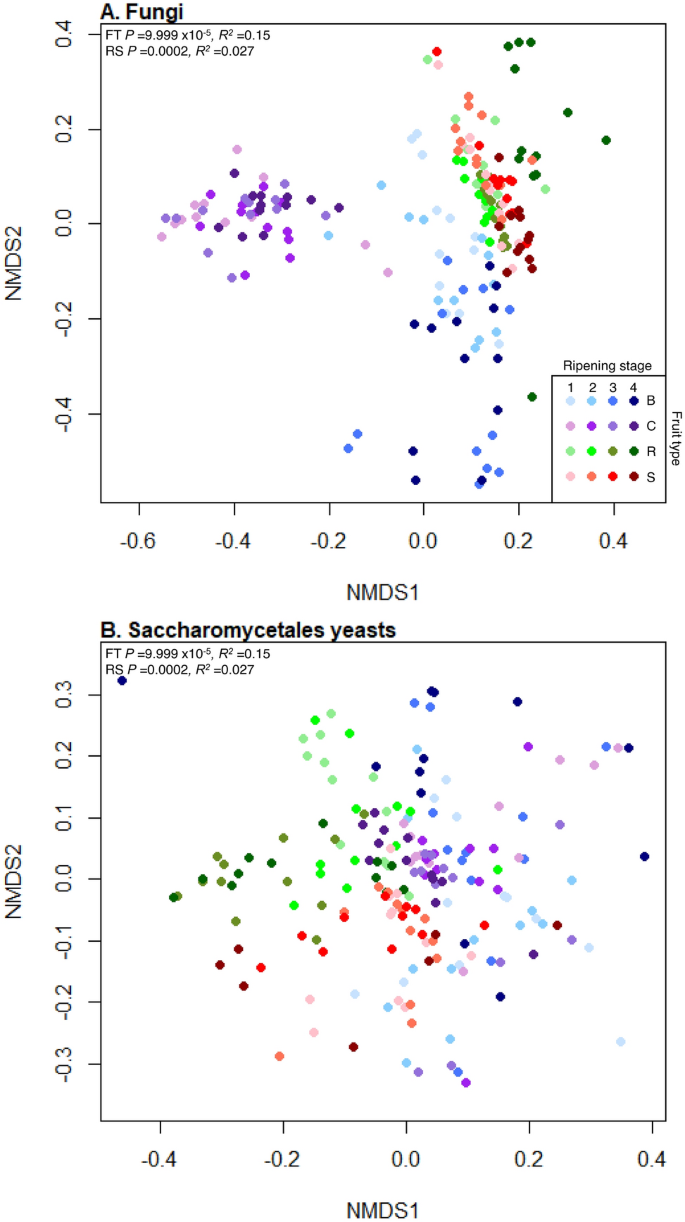
Fruit type and ripening stage also significantly influenced the relative abundances of different fungal phylotypes (PermANOVA, R2 = 0.15, P = 9.999 × 10–5 and R2 = 0.027, P = 0.0002, respectively, Fig. 4A), and the interaction between them was also significant (R2 = 0.018, P = 0.003). Fruit type had approximately 5.6 times greater influence than ripening stage on the relative abundances of fungal phylotypes. Ripening stage significantly influenced the relative abundances of fungal phylotypes present on each fruit separately (blueberry R2 = 0.16, P = 9.999 × 10–5; cherry R2 = 0.061, P = 0.009; raspberry R2 = 0.24, P = 9.999 × 10–5 and strawberry R2 = 0.15, P = 9.999 × 10–5, Fig. S7). There were significant differences in fungal community composition between all fruits and ripening stages (post-hoc pairwise PermANOVAs: P = 9.999 × 10–5, R2 range 0.11–0.57; Supplemental Tables S7, S8).
NMDS plots representing the differential abundances of fungal phylotypes. Nonmetric Multidimensional Scaling (NMDS) plots of abundance Jaccard measures of community dissimilarity of (A) total fungal communities and (B) Saccharomycetales budding yeasts on blueberry (blue), cherry (purple), raspberry (green) and strawberry (red) at four ripening stages (1, unripe/green fruit; 2, de-greening fruit; 3, ripening fruit; and 4, fully ripe/harvest fruit; denoted by shade of colour, lightest shade for green fruit and moving through to darkest shade for fully ripe/harvest). Both total fungal and Saccharomycetales yeasts communities significantly differ in the presences of phylotypes across all fruit types (FT) and ripening stages (RS) by PermANOVA (values shown top left).
Fruit type and ripening stage significantly influenced the relative abundances of Saccharomycetales phylotypes (PermANOVA, R2 = 0.038, P = 9.999 × 10–5 and R2 = 0.024, P = 9.999 × 10–5, respectively, Fig. 4B), with an interaction between the main effects (R2 = 0.016, P = 9.999 × 10–5). Fruit species had approximately 1.6 times greater influence than ripening stage on the relative abundances of phylotypes. Ripening stage significantly affected the relative abundances of Saccharomycetales phylotypes on each fruit separately (blueberry R2 = 0.043, P = 0.004; cherry R2 = 0.64, P = 0.003; raspberry R2 = 0.19, P = 9.999 × 10–5 and strawberry R2 = 0.070, P = 0.0009). There were significant differences in Saccharomycetales community composition between all fruit species and ripening stages (post-hoc pairwise PermANOVA: P = 9.999 × 10–5, R2 range 0.038–0.10; Supplemental Tables S9, S10).
The similarities and differences of fungal phylotypes
The core fruit fungal microbiome
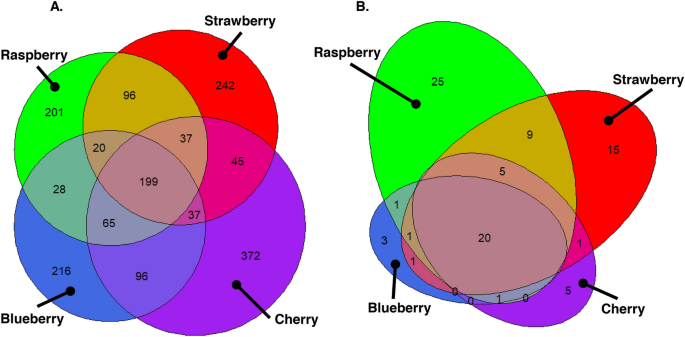
Analyses thus far have focussed on differences in fruit microbiomes, but it is valuable to contrast this with quantifying fruit microbiome similarity. The core fruit fungal microbiome (i.e. those phylotypes present across all fruits) consisted of 199 (11.6%) of the 1712 fungal phylotypes and comprised 97.6% of DNA reads (Table S11). Approximately 12–22% of the 1712 phylotypes were only found associated with specific fruits: 216 with blueberry, 372 with cherry, 201 with raspberry, and 242 with strawberry (Fig. 5A, Table S11). Twenty of the 87 Saccharomycetales phylotypes (23.0%) comprising 81.2% of Saccharomycetales reads were present across all fruit types (Table S11), with 3 unique to blueberry, 5 to cherry, 25 to raspberry and 15 to strawberry (Fig. 5B, Table S12).
The phylotypes that are most differentially abundant
Analyses across all biodiversity metrics show fruit type had a greater effect on fungal communities than maturation stage. Overall, 195 (11.4%) indicator phylotypes (spanning 76 families) had significantly differential abundances between fruit types: 33 phylotypes were significantly overrepresented on blueberry, 70 on cherry, 39 on raspberry and 53 on strawberry (FDR corrected P values ranging from P = 0.011 to P = 0.044). The complete list of significantly differentially overrepresented phylotypes is shown in Table S13 but the two most significantly differentially overrepresented phylotypes on each fruit are listed here: Polyphialoseptoria species and Ramularia (most likely Ramularia endophylla) on blueberry; Exobasidium species and a phylotype from the poorly described order Leotiomycetes on cherry; phylotypes with > 97% homology to Metschnikowia kunwiensis and H. uvarum on raspberry; and phylotypes with > 97% homology to Kalmanozyma fusiformata (Ustilaginaceae smut fungi) and Podosphaera aphanis on strawberry.
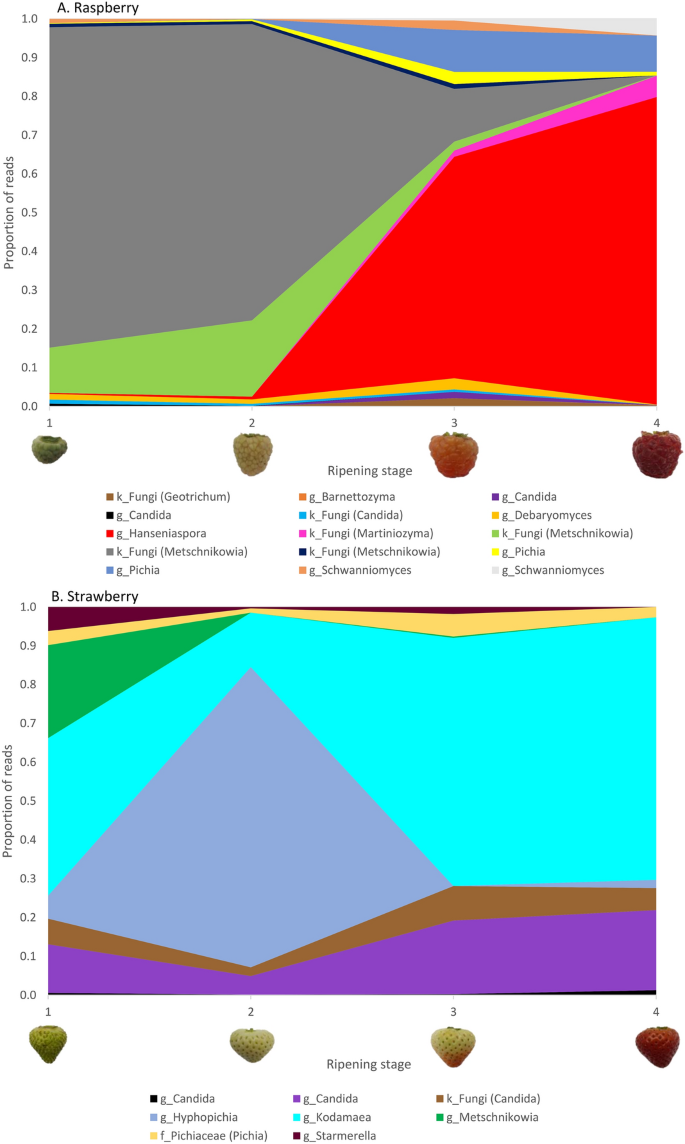
Twenty-four of the 195 indicator phylotypes belonged to the Saccharomycetales budding yeasts (Table S13). There were no Saccharomycetales indicator phylotypes for cherry, and just one for blueberry, a fungal phylotype with > 97% homology to Metschnikowia koreensis. Raspberry had 15 Saccharomycetales indicator phylotypes: three with > 97% homology to the Metschnikowia and, Candida genera, two Pichia and Schwanniomyces, and one each from Hanseniaspora, Barnettozyma, Debaryomyces, Candida, Geotrichum and Martiniozyma. There were eight indicator phylotypes for strawberry; two Candida and one from each of the Metschnikowia, Starmerella, Kodamaea and Hyphopichia genera and the Pichiaceae family, and a phylotype assigned to the no higher level than fungal kingdom (with > 97% homology to deposit from Candida genus). The dynamics of Saccharomycetales yeast indicator phylotypes abundances across maturation for raspberry and strawberry is shown in Fig. 6.
Dynamics of changes in the proportion of budding yeast indicator phylotypes. Mean proportion of reads for the Saccharomycetales budding yeast indicator phylotypes that are significantly overrepresented on (A) raspberry and (B) strawberry (P < 0.04) across the four ripening stages (1, unripe/green fruit; 2, de-greening fruit; 3, ripening fruit; and 4, fully ripe/harvest fruit). Indicator phylotypes are reported to the taxonomic level assigned: lower case letter refers to the taxonomic hierarchy of respective taxa (g = genus; f = family; k = kingdom). Where possible, assignment to genus taxonomic levels is shown in parentheses from matches to deposits in Genbank with > 97% homology identified by manual Blast searches.
Differences of yeast known to be attractive to D. suzukii
Yeast from the Hanseniaspora, Pichia, Saccharomyces, Candida and Metschnikowia genera and their combinations are attractive to D. suzukii27,28,30,31, and phylotypes belonging to these genera were recovered here. The combined relative read abundances of all phylotypes assigned to these genera were significantly different between fruit types and ripening stages (Kruskal–Wallis chi-squared = 60.54, P = 4.51 × 10–13; chi-squared = 10.11, P = 0.018, respectively). Raspberry had the highest relative abundance of yeast genera known to be attractive to D. suzukii (mean ± SE = 21,539 ± 4339) and this was significantly greater than on the other fruits (P < 1.97 × 10–8): cherry (1535 ± 265), strawberry (1651 ± 234) and blueberry (8009 ± 2648). When fruit types were analysed individually, ripening stage had a significant effect on relative read abundance of attractive yeast genera for raspberry only (Kruskal–Wallis chi-squared = 28.70, P = 2.59 × 10–6) where stage 1 and 4 abundances did not significantly differ (mean ± SE = 5682 ± 1522 and 20,826 ± 4711 respectively) but were significantly greater than stage 2 and 3 (2163 ± 538 and 4113 ± 1494 respectively; P < 0.05; Fig. S8).
Various isolates of H. uvarum have consistently been shown to be attractive to D. suzukii27,28,30,31,32. Seven phylotypes were assigned to Hanseniaspora and four of these had > 97% homology to H. uvarum deposits in Genbank using Blast searches, and the relative abundance of these four phylotypes significantly differed between fruit types (Kruskal–Wallis chi-squared = 70.67, df = 3, P = 3.08 × 10–15). Raspberry had the highest relative abundance of H. uvarum (mean ± SE 13,843 ± 3991) but this was not significantly greater (P = 0.080) than abundances on strawberry (426 ± 134) but was significantly greater than blueberry (6 ± 4) and cherry (8 ± 2; P = 1.62 × 10–12, P = 6.00 × 10–9 respectively). Raspberry and strawberry were the only fruits where maturation stage had a significant effect on H. uvarum relative abundance (Kruskal–Wallis chi-squared = 33.40, df = 3, P = 2.66 × 10–7; chi-squared = 12.59, df = 3, P = 0.006), and H. uvarum relative abundance increased as fruits ripened (Fig. S9). This analysis is in line with the indicator phylotype analysis which reported a Hanseniaspora phylotype with > 97% homology to H. uvarum as over-represented on raspberry generally, and especially at later stages (Fig. 6A).
Differences of Botrytis cinerea, known to be repulsive to D. suzukii
The relative read abundances of B. cinerea were significantly different between fruit types and ripening stages (Kruskal–Wallis chi-squared = 73.45, P = 7.80 × 10–16; Kruskal–Wallis chi-squared = 23.81, P = 2.74 × 10–5, respectively). Raspberry had the lowest relative abundance of B. cinerea (mean ± SE = 800 ± 136) and this was significantly lower than strawberry (1994 ± 292) and blueberry (5990 ± 1305) (P < 0.004), but not cherry (3015 ± 1406). Cherry and strawberry had significantly lower reads than blueberry (P < 4.13 × 10–5). When fruit types were analysed individually, ripening stage had a significant effect on relative read abundance of B. cinerea for all fruits (P < 0.003) and reads generally increased during ripening except on cherry where stage 2 had the greatest (Fig. S10).
Correlations with fruit host potential index (HPI) scores
Finally, Bellamy et al.38 generated fruit host potential index (HPI) scores from interactions of D. suzukii with commercial ripe fruit including the fruit species analysed here. The combined relative abundances (i.e. the total number of reads on each fruit across replicates) of yeast phylotypes empirically shown to be attractive to D. suzukii (Hanseniaspora, Pichia, Saccharomyces, Candida and Metschnikowia27,28,29,30,31) across these different ripe fruits at the last sample point are positively correlated with fruit HPI scores (Pearson’s correlation r = 0.38), as are the relative abundance of just H. uvarum (r = 0.62). The relative abundance of B. cinerea was negatively correlated to HPI scores (r = − 0.65) (Fig. S11); however, none of these correlations were significant (P > 0.35) likely due to the low number of comparisons (N = 4 due to just one HPI score per fruit type).
Discussion
Drosophila suzukii is attracted to fungal volatile chemicals (e.g.27); however, little is known about the fungal microbiomes of commercial fruit, with a paucity of information for D. suzukii susceptible fruit. Here we tested the hypothesis that both fruit type and maturation stage have a significant effect on total fruit fungal communities as well as Saccharomycetales yeasts and found strong support for this for all three community biodiversity metrics analysed (numbers, types, and abundances of phylotypes). Raspberry had the greatest relative abundance of yeasts known to be attractive to D. suzukii. Overall, there was a fivefold greater difference in fungal communities between fruit types than maturation stages, showing fruit type was the greater factor defining fruit fungal community assemblage, and cherry had the most distinctive fungal microbiome (Fig. 5). However, there are two main caveats to these conclusions which need consideration. First, we note that fruits matured across different absolute time periods meaning the absolute timing of sampling the various maturation stages differed between fruits, and there is some evidence that other fruit microbiomes can differ through time39,40. Second, fruit fungal communities have been shown to differ by geographic location across hundreds of kilometres6,10,12 and at smaller scales: for example, fungal community dissimilarity increased with distance on grapes from six vineyards separated by a maximum of 35 km11. Therefore, as different fruit sampled in this study were from separate locations up to 19 km apart, it is possible that the variance in fruit-associated fungal microbiomes was also influenced by geographic location. There is support for greater microbiome differentiation by distance from a simple correlation of geographic and community differentiation distances (P = 0.001; Mantle test on distance in km and Jaccard distance); however, distance does not completely explain the variation in fruit microbiomes as cherry and raspberry fungal communities have the greatest dissimilarity (are most separated on NDMS 1 in Fig. 4) but derive from some of the most closely geographically situated sites (Fig. S12). Different fruits would have to be sampled from immediately adjacent sites to completely discount any effect of geographic location on fungal microbiomes. Additionally, while all sites were under conventional management approaches, the precise details of spray programmes are commercially confidential, so it is possible that sites with different fruits were treated with differing spray programmes to control for pest and diseases including fungicides, which may have influenced fungal microbiomes. Some evidence from wine grapes in New Zealand suggests the differences between conventional and biodynamic management only has a small effect on fruit fungal microbiomes14. However, other studies on grapes and pear flowers reported that management practices had a significant effect on total culturable yeasts as well as on community structure13,41. Taken together, the effect of fruit-type detected in this study is likely to be a composite effect of complex interactions of fruit-type × location × management practices and further study on the same fruit species across multiple locations would be necessary to confirm the extent to which fruit species impact microbial communities. Another caveat is that the inference of fungal biodiversity here is derived from the analysis of DNA, and this may not necessarily correlate with phylotypes which are active in communities, and the complementary analyses of RNA may provide an insight into this.
Regardless of the above caveats, these findings show differences in fungal communities on commercial fruit in space and time, and this also holds for species implicated in the attraction of the D. suzukii insect pest. These findings are in line with the few other studies in this field which have shown that fungi differ significantly between apples and blackcurrants4, as well as between sea buckthorn, black chokeberry, red and white currants3. While differences in fungal communities across ripening stages were smaller here, they still changed significantly, especially in the types and abundances of phylotypes, and this agrees with the very limited data from a few other studies evaluating the dynamics of microbiomes as fruit matures8,9,36. However, the temporal dynamics differed between fruit types: numbers remained constant for blueberry but increased with ripening for cherry, and the intermediate ripening stages of raspberry and strawberry had more phylotypes (Fig. 2). Ripening fruit represents a changing habitat which undergoes several physiological changes, including an increase in size and sugar content as well as changes in firmness, colour, and other secondary metabolites which may contribute to fungal community composition. Despite revealing differences in fungal communities between fruit types/sites and maturation stages, there was a large core microbiome which was present across all fruits/sites: this comprised only a fraction of diversity at just 199 of the 1712 fungal phylotypes (Table S11) but was the majority in terms of abundance as it comprised 97.6% of the DNA reads.
Our attempt to quantify changes in absolute fungal cell numbers was only successful for cherry. The total fungal population load per mm2 remained constant across ripening, but there are no other published quantitative DNA based estimates from fruit for comparison. Further optimisation of the levels of added internal standard cells may have allowed quantitative estimates across all fruits; alternatively, adding a synthetic chimeric DNA spike to samples before DNA extraction may be a better strategy42. Using synthetic sequences as an internal standard has the added benefit of this not occurring in environmental samples35. It is also worth noting that including an internal standard in the form of live cells added before DNA extraction assumes that DNA extraction and amplification will be the same across all fungal cells present.
The nature of differences observed for total fungal communities generally held when just Saccharomycetales yeasts were analysed. Specific Saccharomycetales yeast genera which have been empirically shown to be attractive to D. suzukii in field and lab assays27,30 were more prevalent at the raspberry site. Further, the species which has been implicated most in D. suzukii attraction, H. uvarum32, was highly abundant on raspberry. In addition, B. cinerea has been shown to have a repellent effect for D. suzukii34. Of the four fruit sites, raspberry had the lowest amounts of B. cinerea showing an inverse correlation with yeasts attractive to D. suzukii. Raspberry was also the fruit with the greatest host potential index scores for D. suzukii attraction by Bellamy et al.38, and together these observations are in line with the hypothesis that H. uvarum plays a role in D. suzukii attraction to fruit. However, it must be noted that the observed correlation between yeasts shown in other work to be attractive to D. suzukii and the abundances of these yeasts on fruit shown in this study cannot be taken as a causative correlation at this stage. There are other factors like fruit acidity, sugar content and firmness that have been shown to influence in fruit preference of D. suzukii43,44,45. Further work is needed to directly empirically determine the extent to which yeast communities affect D. suzukii preference for fruit. As it stands, these are general correlations, and one may not yet conclude that these abundant yeast phylotypes necessarily drive attraction. The observations here may be compared with a study showing that greater numbers of D. suzukii larvae developed on strawberries than raspberries which where greater than on blueberry; however, this study did not control for fruit associated microbiomes, and factors other than yeast communities may have caused these differing observations46.
Overall, further work is needed to understand if such fruit microbial patterns hold in other locations at other times and whether this correlation with attractive yeast from laboratory and field assays has any underlying basis for causation for D. suzukii fruit susceptibility in the field. If so, this opens the possibility of manipulating fruit microbiomes to deter D. suzukii. Whether fungal species repulsive to D. suzukii species could be ‘seeded’ onto fruits to reduce attractiveness is an intriguing question. Similarly, if this could be combined with traps containing attractive baits situated in and around crops to form a push–pull system to push flies away from fruits and attract them into traps. Although it is unrealistic to use B. cinerea in this way due to its phytopathogenic nature, certain yeast species are known to be repulsive to D. simulans and D. melanogaster30,47,48,49. Van Timmeren et al.50 demonstrated that crop sterilants also impact attractive yeast species growth and reduce D. suzukii larval infestation of fruit. A logical extension of this implies that future data might reveal specific microbes, which are not harmful to fruits or humans and are able to reduce D. suzukii attraction and could therefore be applied for crop protection.
Conclusion
This study demonstrates that for general fungal and more specific Saccharomycetales yeast communities, fruit type or site and maturation stage have a significant impact on fungal diversity, with fruit type/site having a larger effect. This observation also holds for yeast species known to attract D. suzukii, and here these yeasts were most abundant on raspberry30. This knowledge may potentially be applied to better understand what drives D. suzukii susceptibility of different fruit crops at different sites. It is also possible this may inform the engineering of fungal/ yeast communities which could be ‘seeded’ on fruit to reduce the susceptibility of commercial fruit crops to D. suzukii, or to identify ecologically realistic yeast communities for use as potentially attractive phagostimulant baits to control to D. suzukii and reduce the use of chemical pesticides.
Materials and methods
Fruit sampling and processing
All methods, including fruit collections, were performed in accordance with relevant guidelines and the project was conducted under ethics approval CoSREC388 from the University of Lincoln. Based on fruit pigmentation, blueberries, cherries, raspberries, and strawberries were sampled at four developmental stages ranging from unripe (green) to fully ripe (red/purple/navy) (Table S14, Fig. S1) throughout June to September in 2018. Sampling times differed for each fruit type (Table S14). All samples were collected from commercial fruit growers in the United Kingdom southern county of Kent at a maximum distance of 19 km apart; the same sites were revisited at each ripening stage. All fruit were subject to growers’ spray programmes to control pest and diseases. Ten fruits (except blueberries N = 20 as these are smaller) were collected for each species and combined into one sterile bag, and this was replicated six times within each site at each of the four stages for each fruit, totalling 1200 individual and 96 combined fruit samples. Fruits were randomly selected within each field or orchard and were aseptically removed with as little of the stalk or calyx as possible without damaging the fruit. Fruits were briefly inspected for damage before removal with sterile scissors, and fruits were allowed to drop directly into sterile sample bags and thus not handled. Fruits were transported directly to the laboratory where 20 mL of sterile water was added to sample bags. Fruits were then surface-washed repeatedly with this water for 15 s every 5 min for 30 min, after which the contents collected in sterile 50 mL falcon tubes and centrifuged for 30 min at 4500 rpm to collect microbes. No surfactants were used in the surface washing process, as fungi vary in their hydrophobicity this may have affected isolation of certain fungi. The supernatant was reduced to approximately 2 mL, the pellet re-suspended and 1 mL was transferred to microfuge tubes and centrifuged further at 13,000 rpm for 10 min. The supernatant was discarded, and the pellet stored at − 80 °C. After washing, fruit were measured with vernier callipers and surface area estimated using 4πr2.
DNA extraction
Pellets derived from samples were thawed and re-suspended in sterile water, then split into two equal parts. One half of each sample was spiked with 265 live P. cucumerina (Ascomycete: Sordariomycetes) cells determined using a haemocytometer to act as an internal standard. This constituted pairs of samples which were identical other than the spiked P. cucumerina internal standard cells to allow an estimate of absolute cell numbers in the resulting sequence data. Plectosphaerella cucumerina has rarely been reported on the surface of fruits and the isolate used derived from pumpkins in Lincolnshire (UK) and was grown in potato dextrose broth (ThermoFisher Scientific) at 25 °C for 7 days prior to use. Direct cell counts from these fruit samples indicated that 265 cells would represent approximately 0.5% of the community and thus be detectable. DNA was extracted using the DNeasy Blood and Tissue kit (QIAGEN) following the manufacturer’s instructions but with an additional bead beating step before incubation: pellets were resuspended in 750 µL ATL lysis buffer and added to 1 g of sterile glass beads with a 1:1 ratio of < 106 µm: 0.5 mm size (Sigma-Aldrich), then placed in a bead beater (Bead Ruptor 12, Omni international INC) at maximum speed for 5 × 30 s.
Barcode amplification
PCR reactions comprised 15 µL Kapa 2× master mix (Kapa Biosystems), 6 µL of ITS2 forward and reverse primers with Illumina adaptors TS3_KYO251 and ITS452 modified with MiSeq adapters, 7 µL sterile water and 2 µL template DNA. Each batch of PCR reactions included a negative (2 µL sterile water) and positive (S. cerevisiae DNA) control. The PCR cycle parameters were 95 °C for 3 min, 29 cycles of 98 °C for 20 s 64 °C for 20 s 72 °C for 40 s, followed by a final extension time at 72 °C for 5 min. PCR products were separated by electrophoresis using 2% agarose gels containing 10 µL SYBR safe dye™ (Invitrogen) per 100 mL TAE (Tris Acetate-EDTA) buffer (ThermoFisher Scientific). PCR amplicons were sequenced on Illumina MiSeq instruments with a 300PE metric by Eurofins genomics. Raw sequences are deposited on SRA with the following project ID: PRJNA732273.
Bioinformatics analysis
DNA sequences were processed with QIIME 2 (2019.4)53. Sequence quality was evaluated with FastQC54 and reads were trimmed, denoised, paired end merged and ASVs identified with DADA255. ASVs were subsequently clustered with a > 97% genetic identity using vsearch56, and we term ASVs with > 97% identity ‘phylotypes’. Phylotypes assigned to the fungal kingdom were identified using q2-feature-classifier plugin using the unite_ver7dynamic database57; any unassigned phylotypes were subjected to manual Blast searches against the Genbank nucleotide database, and only phylotypes identified as belonging to the fungal kingdom were retained. For non-quantitative analysis, any phylotypes with 100% identity to the P. cucumerina internal standard were removed. Raw sequence counts were subjected to CSS variance‐stabilising normalisation using metagenomSeq and phyloseq R packages58,59,60,61. Recent work indicates that analyses with equal sample depths by rarefication produces the same general patterns as with CSS variance‐stabilising normalisation62, and this is especially important for comparisons of species richness among samples. For the quantitative analysis of fungal communities, samples containing the spiked fungal internal standard (P. cucumerina) were separately processed through the bioinformatics pipeline. Quantitative estimates of phylotype cell counts were calculated by normalising the read number of each phylotype to the number of P. cucumerina reads in that sample, and absolute cell numbers estimated from the knowledge that 265 P. cucumerina cells were added. Phylotype assignments at the species level were estimated by Blast searching the Genbank nucleotide database with representative sequences and reporting hits with > 97% homology. The order Saccharomycetales was analysed by filtering for all phylotypes assigned to Saccharomycetales at the order level.
Statistical analysis
R version 3.6.1 was used for all statistical analyses63. The effect of fruit species and ripening stage on numbers of phylotypes (richness) was assessed using a two-way ANOVA with Tukey HSD for post-hoc pairwise comparisons. A square root transformation was applied where the data did not conform to the assumption of normality as determined by Shapiro-Wilks tests, and Kruskal–Wallis tests applied if transformation did not achieve normality. Omega squared estimates of effect size for two-way ANOVA were calculated with ω2 = dfeffect × (MSeffect − MSerror)/(SStotal + MSerror)64. Shannon’s and Simpson’s diversity indexes were analysed using Kruskal–Wallis tests. Differences in presences or absences of fungal phylotypes and relative abundances of phylotypes were analysed with two‐way full factorial permutational multivariate ANOVA (PermANOVA) using the ‘adonis’ function in the vegan package65 with 10,000 permutations on binary (phylotype presences) and abundance based Jaccard dissimilarity matrices66. Pairwise PermANOVAs were conducted to analyse differences within fruit species and ripening stages where required. For quantitative analysis of fungal communities, the effect of ripening stage on cell numbers was analysed using a Kruskal–Wallis test. Indicator analysis was used to determine fungal phylotypes which were over-represented in the different fruit species with the ‘indicspecies’ package67. ASV abundances were correlated to overall Host Potential Index scores taken from Ref.38 using Pearson’s correlation coefficient. Venn diagrams were created with the ‘eulerr’ package68. Mantel test was used to correlate geographic and community difference using vegan65.
Ethics approval
This project was approved by the University of Lincoln ethics board (CoSREC388).
Data availability
Raw sequences are available on SRA (project ID: PRJNA732273) and the ASV table is provided in the Supplementary Material.
References
-
Ruxton, G. D., Wilkinson, D. M., Schaefer, H. M. & Sherratt, T. N. Why fruit rots: Theoretical support for Janzen’s theory of microbe–macrobe competition. Proc. R. Soc. B Biol. Sci. 281, 20133320. https://doi.org/10.1098/rspb.2013.3320 (2014).
-
Knight, S., Klaere, S., Fedrizzi, B. & Goddard, M. R. Regional microbial signatures positively correlate with differential wine phenotypes: Evidence for a microbial aspect to terroir. Sci. Rep. 5, 1–10. https://doi.org/10.1038/srep14233 (2015).
-
Lukša, J., Vepštaitė-Monstavičė, I., Yurchenko, V., Serva, S. & Servienė, E. High content analysis of sea buckthorn, black chokeberry, red and white currants microbiota—A pilot study. Food Res. Int. 111, 597–606. https://doi.org/10.1016/j.foodres.2018.05.060 (2018).
-
Vepštaitė-Monstavičė, I. et al. Distribution of apple and blackcurrant microbiota in Lithuania and the Czech Republic. Microbiol. Res. 206, 1–8. https://doi.org/10.1016/j.micres.2017.09.004 (2018).
-
Cordero-Bueso, G. et al. Influence of the farming system and vine variety on yeast communities associated with grape berries. Int. J. Food Microbiol. 145, 132–139. https://doi.org/10.1016/J.IJFOODMICRO.2010.11.040 (2011).
-
Gayevskiy, V. & Goddard, M. R. Geographic delineations of yeast communities and populations associated with vines and wines in New Zealand. ISME J. 6, 1281–1290. https://doi.org/10.1038/ismej.2011.195 (2012).
-
Barata, A., Malfeito-Ferreira, M. & Loureiro, V. Changes in sour rotten grape berry microbiota during ripening and wine fermentation. Int. J. Food Microbiol. 154, 152–161. https://doi.org/10.1016/J.IJFOODMICRO.2011.12.029 (2012).
-
Lukša, J. et al. Fungal microbiota of sea buckthorn berries at two ripening stages and volatile profiling of potential biocontrol yeasts. Microorganisms 8, 456. https://doi.org/10.3390/microorganisms8030456 (2020).
-
Abdelfattah, A., Wisniewski, M., Li Destri Nicosia, M. G., Cacciola, S. O. & Schena, L. Metagenomic analysis of fungal diversity on strawberry plants and the effect of management practices on the fungal community structure of aerial organs. PLoS ONE 11, e0160470. https://doi.org/10.1371/journal.pone.0160470 (2016).
-
Bokulich, N. A., Thorngate, J. H., Richardson, P. M. & Mills, D. A. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. 111, E139–E148. https://doi.org/10.1073/PNAS.1317377110 (2014).
-
Miura, T., Sánchez, R., Castañeda, L. E., Godoy, K. & Barbosa, O. Is microbial terroir related to geographic distance between vineyards? Environ. Microbiol. Rep. 9, 742–749. https://doi.org/10.1111/1758-2229.12589 (2017).
-
Taylor, M. W., Tsai, P., Anfang, N., Ross, H. A. & Goddard, M. R. Pyrosequencing reveals regional differences in fruit-associated fungal communities. Environ. Microbiol. 16, 2848–2858. https://doi.org/10.1111/1462-2920.12456 (2014).
-
Martins, G. et al. Influence of the farming system on the epiphytic yeasts and yeast-like fungi colonizing grape berries during the ripening process. Int. J. Food Microbiol. 177, 21–28. https://doi.org/10.1016/J.IJFOODMICRO.2014.02.002 (2014).
-
Morrison-Whittle, P., Lee, S. A. & Goddard, M. R. Fungal communities are differentially affected by conventional and biodynamic agricultural management approaches in vineyard ecosystems. Agric. Ecosyst. Environ. 246, 306–313. https://doi.org/10.1016/j.agee.2017.05.022 (2017).
-
Hamby, K. A. & Becher, P. G. Current knowledge of interactions between Drosophila suzukii and microbes, and their potential utility for pest management. J. Pest. Sci. 89, 621–630. https://doi.org/10.1007/s10340-016-0768-1 (2016).
-
Atallah, J., Teixeira, L., Salazar, R., Zaragoza, G. & Kopp, A. The making of a pest: The evolution of a fruit-penetrating ovipositor in Drosophila suzukii and related species. Proc. R. Soc. B Biol. Sci. 281, 20132840. https://doi.org/10.1098/rspb.2013.2840 (2014).
-
Ioriatti, C. et al. Drosophila suzukii (Diptera: Drosophilidae) contributes to the development of sour rot in grape. J. Econ. Entomol. 111, 283–292. https://doi.org/10.1093/jee/tox292 (2018).
-
Swett, C. L. et al. Characterizing members of the Cladosporium cladosporioides species complex as fruit rot pathogens of red raspberries in the mid-Atlantic and co-occurrence with Drosophila suzukii (spotted wing drosophila). Phytoparasitica 47, 415–428. https://doi.org/10.1007/s12600-019-00734-1 (2019).
-
Lewis, M. T., Koivunen, E. E., Swett, C. L. & Hamby, K. A. Associations between Drosophila suzukii (Diptera: Drosophilidae) and fungi in raspberries. Environ. Entomol. 27, 383–392. https://doi.org/10.1093/ee/nvy167 (2018).
-
Bolda, M. P., Goodhue, R. E. & Zalom, F. G. Spotted wing drosophila: Potential economic impact of a newly established pest. Agric. Resour. Econom. Update 13, 5–8 (2010).
-
Calabria, G., Máca, J., Bächli, G., Serra, L. & Pascual, M. First records of the potential pest species Drosophila suzukii (Diptera: Drosophilidae) in Europe. J. Appl. Entomol. 136, 139–147. https://doi.org/10.1111/j.1439-0418.2010.01583.x (2012).
-
Harris, A. & Shaw, B. First record of Drosophila suzukii (Matsumura) (Diptera, Drosophilidae) in Great Britain. Dipterists Digest 21, 189–192 (2014).
-
Plantamp, C., Estragnat, V., Fellous, S., Desouhant, E. & Gibert, P. Where and what to feed? Differential effects on fecundity and longevity in the invasive Drosophila suzukii. Basic Appl. Ecol. 19, 56–66. https://doi.org/10.1016/j.baae.2016.10.005 (2017).
-
Bellutti, N. et al. Dietary yeast affects preference and performance in Drosophila suzukii. J. Pest. Sci. 91, 651–660. https://doi.org/10.1007/s10340-017-0932-2 (2018).
-
Spitaler, U. et al. Yeast species affects feeding and fitness of Drosophila suzukii adults. J. Pest. Sci. 93, 1295–1309. https://doi.org/10.1007/s10340-020-01266-y (2020).
-
Lewis, M. T. & Hamby, K. A. Differential impacts of yeasts on feeding behavior and development in larval Drosophila suzukii (Diptera:Drosophilidae). Sci. Rep. 9, 13370. https://doi.org/10.1038/s41598-019-48863-1 (2019).
-
Scheidler, N. H., Liu, C., Hamby, K. A., Zalom, F. G. & Syed, Z. Volatile codes: Correlation of olfactory signals and reception in Drosophila-yeast chemical communication. Sci. Rep. 5, 1–13. https://doi.org/10.1038/srep14059 (2015).
-
Noble, R. et al. Improved insecticidal control of spotted wing drosophila (Drosophila suzukii) using yeast and fermented strawberry juice baits. Crop Prot. 125, 104902. https://doi.org/10.1016/J.CROPRO.2019.104902 (2019).
-
Bueno, E. et al. Response of wild spotted wing drosophila (Drosophila suzukii) to microbial volatiles. J. Chem. Ecol. 46, 688–698. https://doi.org/10.1007/s10886-019-01139-4 (2020).
-
Jones, R., Fountain, M. T., Günther, C. S., Eady, P. E. & Goddard, M. R. Separate and combined Hanseniaspora uvarum and Metschnikowia pulcherrima metabolic volatiles are attractive to Drosophila suzukii in the laboratory and field. Sci. Rep. 11, 1201. https://doi.org/10.1038/s41598-020-79691-3 (2021).
-
Lasa, R., Navarro-de-la-Fuente, L., Gschaedler-Mathis, A. C., Kirchmayr, M. R. & Williams, T. Yeast species, strains, and growth media mediate attraction of Drosophila suzukii (Diptera: Drosophilidae). Insects 10, 228. https://doi.org/10.3390/insects10080228 (2019).
-
Hamby, K. A., Hernández, A., Boundy-Mills, K. & Zalom, F. G. Associations of yeasts with spotted-wing Drosophila (Drosophila suzukii; Diptera: Drosophilidae) in cherries and raspberries. Appl. Environ. Microbiol. 78, 4869–4873. https://doi.org/10.1128/AEM.00841-12 (2012).
-
Fountain, M. T. et al. Alimentary microbes of winter-form Drosophila suzukii. Insect Mol. Biol. 27, 383–392. https://doi.org/10.1111/imb.12377 (2018).
-
Cha, D. H. et al. Behavioral evidence for contextual olfactory-mediated avoidance of the ubiquitous phytopathogen Botrytis cinerea by Drosophila suzukii. Insect Sci. 27, 771–779. https://doi.org/10.1111/1744-7917.12691 (2020).
-
Harrison, J. G., John Calder, W., Shuman, B. & Alex Buerkle, C. The quest for absolute abundance: The use of internal standards for DNA-based community ecology. Mol. Ecol. Resour. 21, 30–43. https://doi.org/10.1111/1755-0998.13247 (2021).
-
Carmichael, P. C., Siyoum, N., Chidamba, L. & Korsten, L. Characterization of fungal communities of developmental stages in table grape grown in the northern region of South Africa. J. Appl. Microbiol. 123, 1251–1262. https://doi.org/10.1111/jam.13577 (2017).
-
Morrison-Whittle, P. & Goddard, M. R. From vineyard to winery: A source map of microbial diversity driving wine fermentation. Environ. Microbiol. 20, 75–84. https://doi.org/10.1111/1462-2920.13960 (2018).
-
Bellamy, D. E., Sisterson, M. S. & Walse, S. S. Quantifying host potentials: Indexing postharvest fresh fruits for spotted wing Drosophila, Drosophila suzukii. PLoS ONE 8, e61227. https://doi.org/10.1371/journal.pone.0061227 (2013).
-
Longa, C. M. O. et al. Plant organ and sampling time point determine the taxonomic structure of microbial communities associated to apple plants in the orchard environment. Microbiol. Res. 258, 126991. https://doi.org/10.1016/j.micres.2022.126991 (2022).
-
Shi, X., Chen, Y., Xiao, J., Li, D. & Wang, B. Effects of harvest dates on microbial communities of ice grape skins from Xinjiang of China. Process Biochem. 98, 202–210. https://doi.org/10.1016/j.procbio.2020.08.002 (2020).
-
Schaeffer, R. N. et al. Orchard management and landscape context mediate the pear floral microbiome. Appl. Environ. Microbiol. 87, e00048. https://doi.org/10.1128/AEM.00048-21 (2021).
-
Tkacz, A., Hortala, M. & Poole, P. S. Absolute quantitation of microbiota abundance in environmental samples. Microbiome 6, 110. https://doi.org/10.1186/s40168-018-0491-7 (2018).
-
Burrack, H. J., Fernandez, G. E., Spivey, T. & Kraus, D. A. Variation in selection and utilization of host crops in the field and laboratory by Drosophila suzukii Matsumara (Diptera: Drosophilidae), an invasive frugivore. Pest Manage. Sci. 69, 1173–1180. https://doi.org/10.1002/ps.3489 (2013).
-
Entling, W., Anslinger, S., Jarausch, B., Michl, G. & Hoffmann, C. Berry skin resistance explains oviposition preferences of Drosophila suzukii at the level of grape cultivars and single berries. J. Pest. Sci. 92, 477–484. https://doi.org/10.1007/s10340-018-1040-7 (2019).
-
Ioriatti, C. et al. Drosophila suzukii (Diptera: Drosophilidae) and its potential impact to wine grapes during harvest in two cool climate wine grape production regions. J. Econ. Entomol. 108, 1148–1155. https://doi.org/10.1093/jee/tov042 (2015).
-
Lee, J. C. et al. The susceptibility of small fruits and cherries to the spotted-wing drosophila, Drosophila suzukii. Pest Manage. Sci. 67, 1358–1367. https://doi.org/10.1002/ps.2225 (2011).
-
Buser, C. C., Newcomb, R. D., Gaskett, A. C. & Goddard, M. R. Niche construction initiates the evolution of mutualistic interactions. Ecol. Lett. 17, 1257–1264. https://doi.org/10.1111/ele.12331 (2014).
-
Günther, C. S., Knight, S. J., Jones, R. & Goddard, M. R. Are Drosophila preferences for yeasts stable or contextual? Ecol. Evol. 9, 8075–8086. https://doi.org/10.1002/ece3.5366 (2019).
-
Palanca, L., Gaskett, A. C., Günther, C. S., Newcomb, R. D. & Goddard, M. R. Quantifying variation in the ability of yeasts to attract Drosophila melanogaster. PLoS ONE 8, e75332. https://doi.org/10.1371/journal.pone.0075332 (2013).
-
Van Timmeren, S. et al. Exploring the efficacy and mechanisms of a crop sterilant for reducing infestation by spotted-wing drosophila (Diptera: Drosophilidae). J. Econ. Entomol. 113, 288–298. https://doi.org/10.1093/jee/toz245 (2020).
-
Fujita, S. I., Senda, Y., Nakaguchi, S. & Hashimoto, T. Multiplex PCR using internal transcribed spacer 1 and 2 regions for rapid detection and identification of yeast strains. J. Clin. Microbiol. 39, 3617–3622. https://doi.org/10.1128/jcm.39.10.3617-3622.2001 (2001).
-
White, T. J., Bruns, T., Lee, S. & Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. 18, 315–322 (1990).
-
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. https://doi.org/10.1038/s41587-019-0209-9 (2019).
-
Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc (2010).
-
Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. https://doi.org/10.1038/nmeth.3869 (2016).
-
Rognes, T., Flouri, T., Nichols, B., Quince, C. & Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 4, e2584. https://doi.org/10.7717/peerj.2584 (2016).
-
Bokulich, N. A. et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6, 90. https://doi.org/10.1186/s40168-018-0470-z (2018).
-
McMurdie, P. J. & Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217. https://doi.org/10.1371/journal.pone.0061217 (2013).
-
Muletz Wolz, C. R., Yarwood, S. A., Campbell Grant, E. H., Fleischer, R. C. & Lips, K. R. Effects of host species and environment on the skin microbiome of Plethodontid salamanders. J. Anim. Ecol. 87, 341–353. https://doi.org/10.1111/1365-2656.12726 (2018).
-
Paulson, J. N., Stine, O. C., Bravo, H. C. & Pop, M. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 10, 1200–1202. https://doi.org/10.1038/nmeth.2658 (2013).
-
Weiss, S. et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 5, 27. https://doi.org/10.1186/s40168-017-0237-y (2017).
-
Giraldo-Perez, P., Raw, V., Greven, M. & Goddard, M. R. A small effect of conservation agriculture on soil biodiversity that differs between biological kingdoms and geographic locations. iScience 24, 102280. https://doi.org/10.1016/j.isci.2021.102280 (2021).
-
R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing (2019).
-
Lakens, D. Calculating and reporting effect sizes to facilitate cumulative science: A practical primer for t-tests and ANOVAs. Front. Psychol. 4, 863. https://doi.org/10.3389/fpsyg.2013.00863 (2013).
-
Oksanen, J. et al. Vegan: Community Ecology Package. R Package Version 2.5-6. https://CRAN.R-project.org/package=vegan (2019). Accessed on 24 July 2019.
-
Anderson, M. J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46. https://doi.org/10.1111/J.1442-9993.2001.01070.PP.X (2001).
-
Dufrêne, M. & Legendre, P. Species assemblages and indicator species: The need for a flexible asymmetrical aproach. Ecol. Monogr. 67, 345–366. https://doi.org/10.1890/0012-9615(1997)067[0345:SAAIST]2.0.CO;2 (1997).
-
Larsson, J. Eulerr: Area-proportional Euler and Venn diagrams with ellipses. R Package Version 6.1.0. https://cran.r-project.org/package=eulerr (2020). Accessed on 03 June 2020.
Acknowledgements
We thank the Agriculture and Horticulture Development Board (AHDB) and the University of Lincoln for funding, commercial growers for site access and providing fruit, Geraint Jones for VBA code, and Janet Barlow for proof reading.
Funding
This work was supported by the Agriculture and Horticulture Development Board (contract CP-171) and the University of Lincoln.
Author information
Authors and Affiliations
Contributions
R.J. conducted the experiments, data analyses and wrote the paper; M.G. conceived the study and co-wrote the paper; M.F. conceived the study and commented on drafts of the paper; C.G. helped design experiment and commented on drafts of the paper; N.A. assisted with analyses, Bioinformatics and commented on drafts of the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jones, R., Fountain, M.T., Andreani, N.A. et al. The relative abundances of yeasts attractive to Drosophila suzukii differ between fruit types and are greatest on raspberries. Sci Rep 12, 10382 (2022). https://ift.tt/tsxZmBG
-
Received:
-
Accepted:
-
Published:
-
DOI: https://ift.tt/tsxZmBG
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
"fruit" - Google News
June 21, 2022 at 04:30AM
https://ift.tt/GAzYSMr
The relative abundances of yeasts attractive to Drosophila suzukii differ between fruit types and are greatest on raspberries | Scientific Reports - Nature.com
"fruit" - Google News
https://ift.tt/2HGRMCI
https://ift.tt/vxEM4NZ
Bagikan Berita Ini














0 Response to "The relative abundances of yeasts attractive to Drosophila suzukii differ between fruit types and are greatest on raspberries | Scientific Reports - Nature.com"
Post a Comment